


 |
 |
 |
Patrick AMAR
Bioinfo group, LISN, Univ. Paris Saclay, UMR 9015 CNRS
Synthetic & Systems Biology group, Sys2diag UMR 9005 CNRS, ALCEN
| History |
Silicell Maker has been developed since 2012. The first published reference can be found in the Proceedings of the aDVANCES IN SYSTEMS AND SYNTHETIC BIOLOGY thematic research school, 2015, pp:63-76.
In the PhD memoir of Marc Bouffard (Conception, modélisation et simulation in silico d'un nanosystème biologique artificiel pour le diagnostic médical. Theses, Université Paris Saclay (COmUE), September 2016, in french) one can find a complete description of the NetGate (logic gate finder) and NetBuild (boolean formula to metabolic network) plugins.
| Silicell Maker has been used as the main tool for the research published in Molecular Systems Biology in April 2018. This article, COMPUTER-AIDED BIOCHEMICAL PROGRAMMING OF SYNTHETIC MICROREACTORS AS DIAGNOSTIC DEVICES has been selected to make the cover of the journal. |

|
| Download Silicell Maker |
|
|
|
|
|
|
To install Silicell Maker just put the executable file scmaker (scmaker.exe) in some directory and execute scmaker from this directory. If you want to launch it from everywhere, mention this directory in the PATH variable.
| Quick overview |
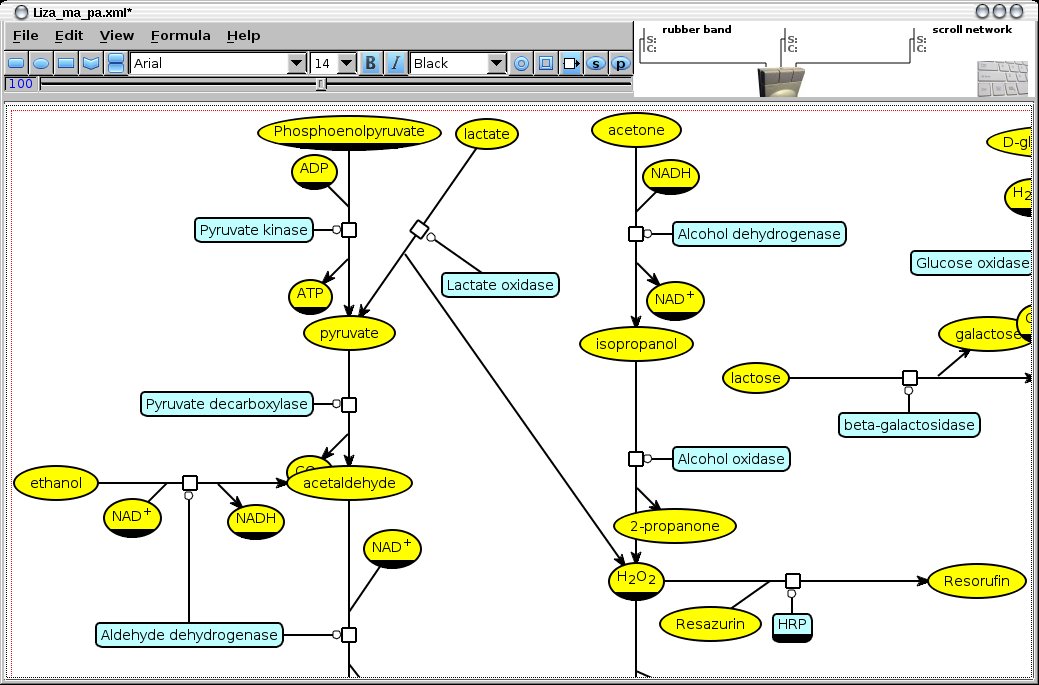
Silicell Maker is a modelling editor / simulator for biochemical network design, with a highly intuitive graphical user interface. It uses the SBGN graphical notation, and support many standard file formats (SBML, Metatool, etc.) Models can be exported to be used by various other modelling tools (HSIM, BIOCHAM, Metatool)
The NetGate and NetBuild plugins are built in Silicell Maker. They allow us to:
Examples of such enzymatic networks used to diagnose diabetic associated diseases can be found in this article.
The logic gates extracted by NetGate are made of sub-networks selected from the input metabolic network. Therefore the choice of this initial network is crucial: it must contains reactions that take the bio-markers as substrates and reactions that produce the revealing metabolites (change of colour, fluorescence, ...).
To help the process, Martin Davy, during his Master 2 internship in the lab, has developed another plugin, BrendaXplorer, that extracts a metabolic network from the BRENDA database, using a supplied list of input and output metabolites. This plugin automatically build the initial metabolic network used by NetGate. Moreover, as the kinetics of the enzymes are known in BRENDA, NetGate can easily find functional logic gates.
Click here to get an HTML version of the on-line user manual.
| Website designed & maintained by Patrick Amar (patrick DOT amar AT lri DOT fr) | 998 hits since 11/01/2017 | |